| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:09 UTC |
|---|
| HMDB ID | HMDB0001388 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | alpha-Linolenic acid |
|---|
| Description | alpha-Linolenic acid (ALA) is a polyunsaturated fatty acid (PUFA). It is a member of the group of essential fatty acids called omega-3 fatty acids. alpha-Linolenic acid, in particular, is not synthesized by mammals and therefore is an essential dietary requirement for all mammals. Certain nuts (English walnuts) and vegetable oils (canola, soybean, flaxseed/linseed, olive) are particularly rich in alpha-linolenic acid. Omega-3 fatty acids get their name based on the location of one of their first double bond. In all omega-3 fatty acids, the first double bond is located between the third and fourth carbon atom counting from the methyl end of the fatty acid (n-3). Although humans and other mammals can synthesize saturated and some monounsaturated fatty acids from carbon groups in carbohydrates and proteins, they lack the enzymes necessary to insert a cis double bond at the n-6 or the n-3 position of a fatty acid. Omega-3 fatty acids like alpha-linolenic acid are important structural components of cell membranes. When incorporated into phospholipids, they affect cell membrane properties such as fluidity, flexibility, permeability, and the activity of membrane-bound enzymes. Omega-3 fatty acids can modulate the expression of a number of genes, including those involved with fatty acid metabolism and inflammation. alpha-Linolenic acid and other omega-3 fatty acids may regulate gene expression by interacting with specific transcription factors, including peroxisome proliferator-activated receptors (PPARs) and liver X receptors (LXRs). alpha-Linolenic acid is found to be associated with isovaleric acidemia, which is an inborn error of metabolism. |
|---|
| Structure | CC\C=C/C\C=C/C\C=C/CCCCCCCC(O)=O InChI=1S/C18H30O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h3-4,6-7,9-10H,2,5,8,11-17H2,1H3,(H,19,20)/b4-3-,7-6-,10-9- |
|---|
| Synonyms | | Value | Source |
|---|
| (9,12,15)-Linolenic acid | ChEBI | | (9Z,12Z,15Z)-Octadecatrienoic acid | ChEBI | | (Z,Z,Z)-9,12,15-Octadecatrienoic acid | ChEBI | | 9,12,15-Octadecatrienoic acid | ChEBI | | 9-cis,12-cis,15-cis-Octadecatrienoic acid | ChEBI | | ALA | ChEBI | | all-cis-9,12,15-Octadecatrienoic acid | ChEBI | | cis,cis,cis-9,12,15-Octadecatrienoic acid | ChEBI | | cis-Delta(9,12,15)-Octadecatrienoic acid | ChEBI | | Linolenic acid | ChEBI | | Linolenate | Kegg | | alpha-Linolenate | Kegg | | (9,12,15)-Linolenate | Generator | | (9Z,12Z,15Z)-Octadecatrienoate | Generator | | (Z,Z,Z)-9,12,15-Octadecatrienoate | Generator | | 9,12,15-Octadecatrienoate | Generator | | 9-cis,12-cis,15-cis-Octadecatrienoate | Generator | | all-cis-9,12,15-Octadecatrienoate | Generator | | cis,cis,cis-9,12,15-Octadecatrienoate | Generator | | cis-delta(9,12,15)-Octadecatrienoate | Generator | | cis-Δ(9,12,15)-octadecatrienoate | Generator | | cis-Δ(9,12,15)-octadecatrienoic acid | Generator | | a-Linolenate | Generator | | a-Linolenic acid | Generator | | Α-linolenate | Generator | | Α-linolenic acid | Generator | | alpha Linolenic acid | MeSH | | cis-9,12,15-Octadecatrienoate | HMDB | | cis-9,12,15-Octadecatrienoic acid | HMDB | | Industrene 120 | HMDB | | FA(18:3(9Z,12Z,15Z)) | HMDB | | FA(18:3n3) | HMDB |
|
|---|
| Chemical Formula | C18H30O2 |
|---|
| Average Molecular Weight | 278.4296 |
|---|
| Monoisotopic Molecular Weight | 278.224580204 |
|---|
| IUPAC Name | (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid |
|---|
| Traditional Name | α linolenic acid |
|---|
| CAS Registry Number | 463-40-1 |
|---|
| SMILES | CC\C=C/C\C=C/C\C=C/CCCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H30O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h3-4,6-7,9-10H,2,5,8,11-17H2,1H3,(H,19,20)/b4-3-,7-6-,10-9- |
|---|
| InChI Key | DTOSIQBPPRVQHS-PDBXOOCHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Lineolic acids and derivatives |
|---|
| Direct Parent | Lineolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Octadecanoid
- Long-chain fatty acid
- Fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | Naturally occurring processBiological processBiochemical pathwayMetabolic pathway- Alpha Linolenic Acid and Linoleic Acid Metabolism (HMDB: HMDB0001388)
- Fatty Acid Metabolism (HMDB: HMDB0001388)
- Triacylglycerol Degradation TG(16:0/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012474)
- Triacylglycerol Degradation TG(16:0/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012486)
- Triacylglycerol Degradation TG(16:0/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012498)
- Triacylglycerol Degradation TG(16:0/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012510)
- Triacylglycerol Degradation TG(16:0/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012522)
- Triacylglycerol Degradation TG(16:0/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012534)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0012540)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0012541)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0012542)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0012543)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0012544)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0012545)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012546)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0012547)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0012548)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0012549)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0012550)
- Triacylglycerol Degradation TG(16:0/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0012551)
- Triacylglycerol Degradation TG(16:0/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012558)
- Triacylglycerol Degradation TG(16:0/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012570)
- Triacylglycerol Degradation TG(16:0/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012582)
- Triacylglycerol Degradation TG(16:0/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012594)
- Triacylglycerol Degradation TG(16:0/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012606)
- Triacylglycerol Degradation TG(18:0/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012618)
- Triacylglycerol Degradation TG(18:0/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012630)
- Triacylglycerol Degradation TG(18:0/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012642)
- Triacylglycerol Degradation TG(18:0/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012654)
- Triacylglycerol Degradation TG(18:0/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012666)
- Triacylglycerol Degradation TG(18:0/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012678)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0012684)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0012685)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0012686)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0012687)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0012688)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0012689)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012690)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0012691)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0012692)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0012693)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0012694)
- Triacylglycerol Degradation TG(18:0/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0012695)
- Triacylglycerol Degradation TG(18:0/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012702)
- Triacylglycerol Degradation TG(18:0/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012714)
- Triacylglycerol Degradation TG(18:0/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012726)
- Triacylglycerol Degradation TG(18:0/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012738)
- Triacylglycerol Degradation TG(18:0/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012750)
- Triacylglycerol Degradation TG(18:1(9Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012762)
- Triacylglycerol Degradation TG(18:1(9Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012774)
- Triacylglycerol Degradation TG(18:1(9Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012786)
- Triacylglycerol Degradation TG(18:1(9Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012798)
- Triacylglycerol Degradation TG(18:1(9Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012810)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012822)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0012828)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0012829)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0012830)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0012831)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0012832)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0012833)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012834)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0012835)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0012836)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0012837)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0012838)
- Triacylglycerol Degradation TG(18:1(9Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0012839)
- Triacylglycerol Degradation TG(18:1(9Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012846)
- Triacylglycerol Degradation TG(18:1(9Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012858)
- Triacylglycerol Degradation TG(18:1(9Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012870)
- Triacylglycerol Degradation TG(18:1(9Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012882)
- Triacylglycerol Degradation TG(18:1(9Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012894)
- Triacylglycerol Degradation TG(18:1(11Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012906)
- Triacylglycerol Degradation TG(18:1(11Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012918)
- Triacylglycerol Degradation TG(18:1(11Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012930)
- Triacylglycerol Degradation TG(18:1(11Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012942)
- Triacylglycerol Degradation TG(18:1(11Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012954)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012966)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0012972)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0012973)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0012974)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0012975)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0012976)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0012977)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0012978)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0012979)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0012980)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0012981)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0012982)
- Triacylglycerol Degradation TG(18:1(11Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0012983)
- Triacylglycerol Degradation TG(18:1(11Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0012990)
- Triacylglycerol Degradation TG(18:1(11Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013002)
- Triacylglycerol Degradation TG(18:1(11Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013014)
- Triacylglycerol Degradation TG(18:1(11Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013026)
- Triacylglycerol Degradation TG(18:1(11Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013038)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013050)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013062)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013074)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013086)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013098)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013110)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013116)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013117)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013118)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013119)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013120)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013121)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013122)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013123)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013124)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013125)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013126)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013127)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013134)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013146)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013158)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013170)
- Triacylglycerol Degradation TG(18:2(9Z,12Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013182)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013194)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013206)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013218)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013230)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013242)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013254)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013260)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013261)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013262)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013263)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013264)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013265)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013266)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013267)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013268)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013269)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013270)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013271)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013278)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013290)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013302)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013314)
- Triacylglycerol Degradation TG(18:3(6Z,9Z,12Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013326)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/16:0) (PathBank: SMP0013332)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:0) (PathBank: SMP0013333)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:1(9Z)) (PathBank: SMP0013334)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:1(11Z)) (PathBank: SMP0013335)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:2(9Z,12Z)) (PathBank: SMP0013336)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:3(6Z,9Z,12Z)) (PathBank: SMP0013337)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013338)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/20:0) (PathBank: SMP0013339)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/20:1(11Z)) (PathBank: SMP0013340)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/20:1(13Z)) (PathBank: SMP0013341)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/22:0) (PathBank: SMP0013342)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/16:0/22:1(13Z)) (PathBank: SMP0013343)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/16:0) (PathBank: SMP0013344)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:0) (PathBank: SMP0013345)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:1(9Z)) (PathBank: SMP0013346)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:1(11Z)) (PathBank: SMP0013347)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:2(9Z,12Z)) (PathBank: SMP0013348)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:3(6Z,9Z,12Z)) (PathBank: SMP0013349)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013350)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/20:0) (PathBank: SMP0013351)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/20:1(11Z)) (PathBank: SMP0013352)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/20:1(13Z)) (PathBank: SMP0013353)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/22:0) (PathBank: SMP0013354)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:0/22:1(13Z)) (PathBank: SMP0013355)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/16:0) (PathBank: SMP0013356)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:0) (PathBank: SMP0013357)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:1(9Z)) (PathBank: SMP0013358)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:1(11Z)) (PathBank: SMP0013359)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:2(9Z,12Z)) (PathBank: SMP0013360)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013361)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/20:0) (PathBank: SMP0013362)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/20:1(11Z)) (PathBank: SMP0013363)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/20:1(13Z)) (PathBank: SMP0013364)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/22:0) (PathBank: SMP0013365)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/22:1(13Z)) (PathBank: SMP0013366)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/16:0) (PathBank: SMP0013367)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:0) (PathBank: SMP0013368)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:1(9Z)) (PathBank: SMP0013369)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:1(11Z)) (PathBank: SMP0013370)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:2(9Z,12Z)) (PathBank: SMP0013371)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013372)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/20:0) (PathBank: SMP0013373)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/20:1(11Z)) (PathBank: SMP0013374)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/20:1(13Z)) (PathBank: SMP0013375)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/22:0) (PathBank: SMP0013376)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/22:1(13Z)) (PathBank: SMP0013377)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/16:0) (PathBank: SMP0013378)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:0) (PathBank: SMP0013379)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:1(9Z)) (PathBank: SMP0013380)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:1(11Z)) (PathBank: SMP0013381)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:2(9Z,12Z)) (PathBank: SMP0013382)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013383)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/20:0) (PathBank: SMP0013384)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/20:1(11Z)) (PathBank: SMP0013385)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/20:1(13Z)) (PathBank: SMP0013386)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/22:0) (PathBank: SMP0013387)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/22:1(13Z)) (PathBank: SMP0013388)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/16:0) (PathBank: SMP0013389)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:0) (PathBank: SMP0013390)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:1(9Z)) (PathBank: SMP0013391)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:1(11Z)) (PathBank: SMP0013392)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:2(9Z,12Z)) (PathBank: SMP0013393)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013394)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013395)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/20:0) (PathBank: SMP0013396)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/20:1(11Z)) (PathBank: SMP0013397)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/20:1(13Z)) (PathBank: SMP0013398)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/22:0) (PathBank: SMP0013399)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)/22:1(13Z)) (PathBank: SMP0013400)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013401)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013402)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013403)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013404)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013405)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013406)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013407)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013408)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013409)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013410)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013411)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013412)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/16:0) (PathBank: SMP0013413)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:0) (PathBank: SMP0013414)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:1(9Z)) (PathBank: SMP0013415)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:1(11Z)) (PathBank: SMP0013416)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:2(9Z,12Z)) (PathBank: SMP0013417)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:3(6Z,9Z,12Z)) (PathBank: SMP0013418)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013419)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/20:0) (PathBank: SMP0013420)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/20:1(11Z)) (PathBank: SMP0013421)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/20:1(13Z)) (PathBank: SMP0013422)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/22:0) (PathBank: SMP0013423)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:0/22:1(13Z)) (PathBank: SMP0013424)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/16:0) (PathBank: SMP0013425)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:0) (PathBank: SMP0013426)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:1(9Z)) (PathBank: SMP0013427)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:1(11Z)) (PathBank: SMP0013428)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:2(9Z,12Z)) (PathBank: SMP0013429)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013430)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013431)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/20:0) (PathBank: SMP0013432)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/20:1(11Z)) (PathBank: SMP0013433)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/20:1(13Z)) (PathBank: SMP0013434)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/22:0) (PathBank: SMP0013435)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(11Z)/22:1(13Z)) (PathBank: SMP0013436)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/16:0) (PathBank: SMP0013437)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:0) (PathBank: SMP0013438)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:1(9Z)) (PathBank: SMP0013439)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:1(11Z)) (PathBank: SMP0013440)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:2(9Z,12Z)) (PathBank: SMP0013441)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013442)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013443)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/20:0) (PathBank: SMP0013444)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/20:1(11Z)) (PathBank: SMP0013445)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/20:1(13Z)) (PathBank: SMP0013446)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/22:0) (PathBank: SMP0013447)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/20:1(13Z)/22:1(13Z)) (PathBank: SMP0013448)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/16:0) (PathBank: SMP0013449)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:0) (PathBank: SMP0013450)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:1(9Z)) (PathBank: SMP0013451)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:1(11Z)) (PathBank: SMP0013452)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:2(9Z,12Z)) (PathBank: SMP0013453)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:3(6Z,9Z,12Z)) (PathBank: SMP0013454)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013455)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/20:0) (PathBank: SMP0013456)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/20:1(11Z)) (PathBank: SMP0013457)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/20:1(13Z)) (PathBank: SMP0013458)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/22:0) (PathBank: SMP0013459)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:0/22:1(13Z)) (PathBank: SMP0013460)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/16:0) (PathBank: SMP0013461)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:0) (PathBank: SMP0013462)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:1(9Z)) (PathBank: SMP0013463)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:1(11Z)) (PathBank: SMP0013464)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:2(9Z,12Z)) (PathBank: SMP0013465)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013466)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013467)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/20:0) (PathBank: SMP0013468)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/20:1(11Z)) (PathBank: SMP0013469)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/20:1(13Z)) (PathBank: SMP0013470)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/22:0) (PathBank: SMP0013471)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/22:1(13Z)/22:1(13Z)) (PathBank: SMP0013472)
- Triacylglycerol Degradation TG(20:0/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013479)
- Triacylglycerol Degradation TG(20:0/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013491)
- Triacylglycerol Degradation TG(20:0/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013503)
- Triacylglycerol Degradation TG(20:0/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013515)
- Triacylglycerol Degradation TG(20:0/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013527)
- Triacylglycerol Degradation TG(20:0/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013539)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013545)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013546)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013547)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013548)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013549)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013550)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013551)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013552)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013553)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013554)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013555)
- Triacylglycerol Degradation TG(20:0/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013556)
- Triacylglycerol Degradation TG(20:0/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013563)
- Triacylglycerol Degradation TG(20:0/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013575)
- Triacylglycerol Degradation TG(20:0/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013587)
- Triacylglycerol Degradation TG(20:0/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013599)
- Triacylglycerol Degradation TG(20:0/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013611)
- Triacylglycerol Degradation TG(20:1(11Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013623)
- Triacylglycerol Degradation TG(20:1(11Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013635)
- Triacylglycerol Degradation TG(20:1(11Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013647)
- Triacylglycerol Degradation TG(20:1(11Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013659)
- Triacylglycerol Degradation TG(20:1(11Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013671)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013683)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013689)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013690)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013691)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013692)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013693)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013694)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013695)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013696)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013697)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013698)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013699)
- Triacylglycerol Degradation TG(20:1(11Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013700)
- Triacylglycerol Degradation TG(20:1(11Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013707)
- Triacylglycerol Degradation TG(20:1(11Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013719)
- Triacylglycerol Degradation TG(20:1(11Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013731)
- Triacylglycerol Degradation TG(20:1(11Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013743)
- Triacylglycerol Degradation TG(20:1(11Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013755)
- Triacylglycerol Degradation TG(20:1(13Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013768)
- Triacylglycerol Degradation TG(20:1(13Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013780)
- Triacylglycerol Degradation TG(20:1(13Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013792)
- Triacylglycerol Degradation TG(20:1(13Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013804)
- Triacylglycerol Degradation TG(20:1(13Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013816)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013828)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013834)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013835)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013836)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013837)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013838)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013839)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013840)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013841)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013842)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013843)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013844)
- Triacylglycerol Degradation TG(20:1(13Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013845)
- Triacylglycerol Degradation TG(20:1(13Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013852)
- Triacylglycerol Degradation TG(20:1(13Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013864)
- Triacylglycerol Degradation TG(20:1(13Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013876)
- Triacylglycerol Degradation TG(20:1(13Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013888)
- Triacylglycerol Degradation TG(20:1(13Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013901)
- Triacylglycerol Degradation TG(22:0/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013913)
- Triacylglycerol Degradation TG(22:0/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013925)
- Triacylglycerol Degradation TG(22:0/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013937)
- Triacylglycerol Degradation TG(22:0/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013949)
- Triacylglycerol Degradation TG(22:0/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013961)
- Triacylglycerol Degradation TG(22:0/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013973)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0013979)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0013980)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0013981)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0013982)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0013983)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0013984)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0013985)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0013986)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0013987)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0013988)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0013989)
- Triacylglycerol Degradation TG(22:0/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0013990)
- Triacylglycerol Degradation TG(22:0/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0013997)
- Triacylglycerol Degradation TG(22:0/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014009)
- Triacylglycerol Degradation TG(22:0/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014023)
- Triacylglycerol Degradation TG(22:0/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0014035)
- Triacylglycerol Degradation TG(22:0/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014047)
- Triacylglycerol Degradation TG(22:1(13Z)/16:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0014059)
- Triacylglycerol Degradation TG(22:1(13Z)/18:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0014071)
- Triacylglycerol Degradation TG(22:1(13Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014083)
- Triacylglycerol Degradation TG(22:1(13Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014095)
- Triacylglycerol Degradation TG(22:1(13Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014107)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(6Z,9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014119)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/16:0) (PathBank: SMP0014125)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:0) (PathBank: SMP0014126)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:1(9Z)) (PathBank: SMP0014127)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:1(11Z)) (PathBank: SMP0014128)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:2(9Z,12Z)) (PathBank: SMP0014129)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:3(6Z,9Z,12Z)) (PathBank: SMP0014130)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014131)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/20:0) (PathBank: SMP0014132)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/20:1(11Z)) (PathBank: SMP0014133)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/20:1(13Z)) (PathBank: SMP0014134)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/22:0) (PathBank: SMP0014135)
- Triacylglycerol Degradation TG(22:1(13Z)/18:3(9Z,12Z,15Z)/22:1(13Z)) (PathBank: SMP0014136)
- Triacylglycerol Degradation TG(22:1(13Z)/20:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0014143)
- Triacylglycerol Degradation TG(22:1(13Z)/20:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014155)
- Triacylglycerol Degradation TG(22:1(13Z)/20:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014168)
- Triacylglycerol Degradation TG(22:1(13Z)/22:0/18:3(9Z,12Z,15Z)) (PathBank: SMP0014180)
- Triacylglycerol Degradation TG(22:1(13Z)/22:1(13Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014192)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(9Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014199)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:1(11Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014200)
- Triacylglycerol Degradation TG(18:3(9Z,12Z,15Z)/18:2(9Z,12Z)/18:3(9Z,12Z,15Z)) (PathBank: SMP0014201)
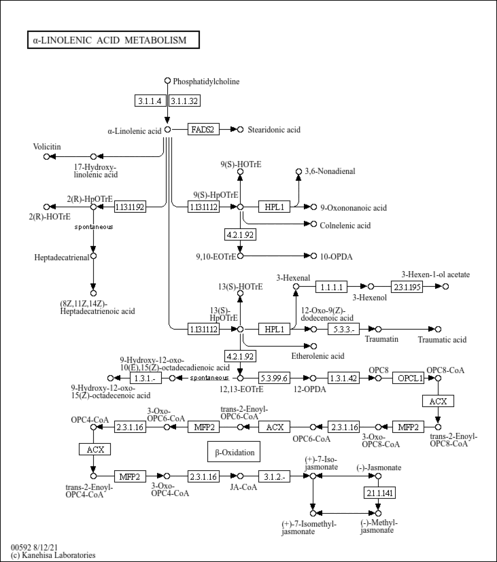
- Alpha-Linolenic Acid Metabolism (PathBank: SMP0121229)
- Lipid metabolism pathway (HMDB: HMDB0001388)
Signaling pathway Chemical reactionCellular processBiochemical process |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | -16.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | 6.46 | SANGSTER (1993) |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (1 TMS) | splash10-052f-7900000000-8765cf82603feb1b448f | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (1 TMS) | splash10-004i-9300000000-302519a616eaed5fcc59 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-MS (1 TMS) | splash10-005c-9800000000-2d44b3c70e2992b9a812 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-EI-TOF (Non-derivatized) | splash10-052f-7900000000-8765cf82603feb1b448f | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-EI-TOF (Non-derivatized) | splash10-004i-9300000000-302519a616eaed5fcc59 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-MS (Non-derivatized) | splash10-005c-9800000000-2d44b3c70e2992b9a812 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - alpha-Linolenic acid GC-EI-TOF (Non-derivatized) | splash10-052f-5900000000-28710ea35f196c595e03 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - alpha-Linolenic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-052e-6950000000-27037263e74c8119ead0 | 2017-08-28 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - alpha-Linolenic acid GC-MS (1 TMS) - 70eV, Positive | splash10-00ds-6931000000-657fa0cc66a206a36409 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - alpha-Linolenic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid Quattro_QQQ 10V, N/A-QTOF (Annotated) | splash10-004i-0190000000-aee97e9ea2c7f0783adf | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid Quattro_QQQ 25V, N/A-QTOF (Annotated) | splash10-05nb-9500000000-fb92aefc701027239359 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid Quattro_QQQ 40V, N/A-QTOF (Annotated) | splash10-0aru-9100000000-a5dddfe6f629d1371fac | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0090000000-2b7bea03a685454dd135 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid ESI-TOF , Negative-QTOF | splash10-004i-0090000000-53ddfaed6ce13c37d957 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid ESI-TOF 20V, Negative-QTOF | splash10-004i-0090000000-e5350a66361bc63d1071 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid ESI-TOF 30V, Negative-QTOF | splash10-004i-0090000000-5731aa5e301022c06813 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0091010000-6922411e48d747e592b5 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-IT , negative-QTOF | splash10-001i-0090000000-a18b573ad888d048efa6 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-TOF , negative-QTOF | splash10-004i-0090000000-2b7bea03a685454dd135 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-TOF , negative-QTOF | splash10-004i-0090000000-e5350a66361bc63d1071 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-TOF , negative-QTOF | splash10-004i-0090000000-5731aa5e301022c06813 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-QTOF 10V, positive-QTOF | splash10-004i-0090000000-1d391027db3704d71cb9 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-QTOF 20V, positive-QTOF | splash10-00lr-9210000000-41acd8a55f52afec6d79 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid LC-ESI-QTOF 40V, positive-QTOF | splash10-014i-9000000000-a976b4a80a07eb2c4641 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid 40V, Positive-QTOF | splash10-0a4i-9100000000-497eddf5d738a45f83fb | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid 10V, Negative-QTOF | splash10-004i-0090000000-0c00ca98901526cb2e26 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid 30V, Negative-QTOF | splash10-004i-1090000000-35ffce9343fe0d15ea0d | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - alpha-Linolenic acid 30V, Negative-QTOF | splash10-004i-0090000000-373584fbab950d6676cc | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 10V, Positive-QTOF | splash10-01t9-0090000000-ae1f482177d0e208165e | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 20V, Positive-QTOF | splash10-00o0-5690000000-20804c8382f14ff0ccf4 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 40V, Positive-QTOF | splash10-014l-8930000000-9d5e342b1351adc71c20 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 10V, Negative-QTOF | splash10-004i-0090000000-4d3e8d1180800a7b4ed5 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 20V, Negative-QTOF | splash10-0059-1090000000-67ab14c068a142dd3c30 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - alpha-Linolenic acid 40V, Negative-QTOF | splash10-0a4i-9230000000-403c610d63380e63109a | 2017-07-26 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
|
|---|
| Disease References | | Hypertension |
|---|
- Wang S, Ma A, Song S, Quan Q, Zhao X, Zheng X: Fasting serum free fatty acid composition, waist/hip ratio and insulin activity in essential hypertensive patients. Hypertens Res. 2008 Apr;31(4):623-32. doi: 10.1291/hypres.31.623. [PubMed:18633173 ]
| | Essential hypertension |
|---|
- Wang S, Ma A, Song S, Quan Q, Zhao X, Zheng X: Fasting serum free fatty acid composition, waist/hip ratio and insulin activity in essential hypertensive patients. Hypertens Res. 2008 Apr;31(4):623-32. doi: 10.1291/hypres.31.623. [PubMed:18633173 ]
| | Major depressive disorder |
|---|
- Sublette ME, Segal-Isaacson CJ, Cooper TB, Fekri S, Vanegas N, Galfalvy HC, Oquendo MA, Mann JJ: Validation of a food frequency questionnaire to assess intake of n-3 polyunsaturated fatty acids in subjects with and without major depressive disorder. J Am Diet Assoc. 2011 Jan;111(1):117-123.e1-2. doi: 10.1016/j.jada.2010.10.007. [PubMed:21185973 ]
| | Isovaleric acidemia |
|---|
- Dercksen M, Kulik W, Mienie LJ, Reinecke CJ, Wanders RJ, Duran M: Polyunsaturated fatty acid status in treated isovaleric acidemia patients. Eur J Clin Nutr. 2016 Oct;70(10):1123-1126. doi: 10.1038/ejcn.2016.100. Epub 2016 Jun 22. [PubMed:27329611 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Thyroid cancer |
|---|
- Kim KM, Jung BH, Lho DS, Chung WY, Paeng KJ, Chung BC: Alteration of urinary profiles of endogenous steroids and polyunsaturated fatty acids in thyroid cancer. Cancer Lett. 2003 Dec 30;202(2):173-9. [PubMed:14643447 ]
|
|
|---|
| General References | - Richieri GV, Ogata RT, Kleinfeld AM: Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J Biol Chem. 1994 Sep 30;269(39):23918-30. [PubMed:7929039 ]
- Bajaj M, Suraamornkul S, Romanelli A, Cline GW, Mandarino LJ, Shulman GI, DeFronzo RA: Effect of a sustained reduction in plasma free fatty acid concentration on intramuscular long-chain fatty Acyl-CoAs and insulin action in type 2 diabetic patients. Diabetes. 2005 Nov;54(11):3148-53. [PubMed:16249438 ]
- Christensen JH, Fabrin K, Borup K, Barber N, Poulsen J: Prostate tissue and leukocyte levels of n-3 polyunsaturated fatty acids in men with benign prostate hyperplasia or prostate cancer. BJU Int. 2006 Feb;97(2):270-3. [PubMed:16430627 ]
- Attar-Bashi NM, Frauman AG, Sinclair AJ: Alpha-linolenic acid and the risk of prostate cancer. What is the evidence? J Urol. 2004 Apr;171(4):1402-7. [PubMed:15017185 ]
- Rastogi SK, Singh J: Effect of chemical penetration enhancer and iontophoresis on the in vitro percutaneous absorption enhancement of insulin through porcine epidermis. Pharm Dev Technol. 2005;10(1):97-104. [PubMed:15776817 ]
- Allman MA, Pena MM, Pang D: Supplementation with flaxseed oil versus sunflowerseed oil in healthy young men consuming a low fat diet: effects on platelet composition and function. Eur J Clin Nutr. 1995 Mar;49(3):169-78. [PubMed:7774533 ]
- Fokkema MR, Brouwer DA, Hasperhoven MB, Martini IA, Muskiet FA: Short-term supplementation of low-dose gamma-linolenic acid (GLA), alpha-linolenic acid (ALA), or GLA plus ALA does not augment LCP omega 3 status of Dutch vegans to an appreciable extent. Prostaglandins Leukot Essent Fatty Acids. 2000 Nov;63(5):287-92. [PubMed:11090255 ]
- Becker CC, Lund P, Holmer G, Jensen H, Sandstrom B: Effects of butter oil blends with increased concentrations of stearic, oleic and linolenic acid on blood lipids in young adults. Eur J Clin Nutr. 1999 Jul;53(7):535-41. [PubMed:10452408 ]
- Jones DB, Scaretto L, Carter R, Mann JI: Glucose, insulin and platelet fatty acids following myocardial infarction: an association with infarct size. Diabete Metab. 1987 Jul-Aug;13(4):463-6. [PubMed:3315767 ]
- Crastes de Paulet A, Babin F, Billeaud C, Bougle D, Sarda P, Mendy F: [Biological effects on premature neonates of a milk formula enriched with alpha-linolenic acid: a multicenter study]. Bull Acad Natl Med. 1994 Feb;178(2):267-73; discussion 273-8. [PubMed:7913655 ]
- Li D, Sinclair A, Wilson A, Nakkote S, Kelly F, Abedin L, Mann N, Turner A: Effect of dietary alpha-linolenic acid on thrombotic risk factors in vegetarian men. Am J Clin Nutr. 1999 May;69(5):872-82. [PubMed:10232625 ]
- Bhatia KS, Singh J: Effect of linolenic acid/ethanol or limonene/ethanol and iontophoresis on the in vitro percutaneous absorption of LHRH and ultrastructure of human epidermis. Int J Pharm. 1999 Apr 15;180(2):235-50. [PubMed:10370194 ]
- Baylin A, Kabagambe EK, Ascherio A, Spiegelman D, Campos H: Adipose tissue alpha-linolenic acid and nonfatal acute myocardial infarction in Costa Rica. Circulation. 2003 Apr 1;107(12):1586-91. Epub 2003 Mar 10. [PubMed:12668490 ]
- Williard DE, Nwankwo JO, Kaduce TL, Harmon SD, Irons M, Moser HW, Raymond GV, Spector AA: Identification of a fatty acid delta6-desaturase deficiency in human skin fibroblasts. J Lipid Res. 2001 Apr;42(4):501-8. [PubMed:11290821 ]
- Campbell FM, Gordon MJ, Dutta-Roy AK: Preferential uptake of long chain polyunsaturated fatty acids by isolated human placental membranes. Mol Cell Biochem. 1996 Feb 9;155(1):77-83. [PubMed:8717442 ]
- Cunnane SC, Hamadeh MJ, Liede AC, Thompson LU, Wolever TM, Jenkins DJ: Nutritional attributes of traditional flaxseed in healthy young adults. Am J Clin Nutr. 1995 Jan;61(1):62-8. [PubMed:7825540 ]
- Connor WE: Importance of n-3 fatty acids in health and disease. Am J Clin Nutr. 2000 Jan;71(1 Suppl):171S-5S. [PubMed:10617967 ]
- Lauritzen I, Blondeau N, Heurteaux C, Widmann C, Romey G, Lazdunski M: Polyunsaturated fatty acids are potent neuroprotectors. EMBO J. 2000 Apr 17;19(8):1784-93. [PubMed:10775263 ]
- Cho E, Hung S, Willett WC, Spiegelman D, Rimm EB, Seddon JM, Colditz GA, Hankinson SE: Prospective study of dietary fat and the risk of age-related macular degeneration. Am J Clin Nutr. 2001 Feb;73(2):209-18. [PubMed:11157315 ]
- Kris-Etherton PM, Harris WS, Appel LJ: Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 2002 Nov 19;106(21):2747-57. [PubMed:12438303 ]
- Brouwer IA, Katan MB, Zock PL: Dietary alpha-linolenic acid is associated with reduced risk of fatal coronary heart disease, but increased prostate cancer risk: a meta-analysis. J Nutr. 2004 Apr;134(4):919-22. [PubMed:15051847 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|