| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:09 UTC |
|---|
| HMDB ID | HMDB0001375 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Hydroxy-3-methylglutaryl-CoA |
|---|
| Description | 3-Hydroxy-3-methylglutaryl-CoA (HMG-CoA) (CAS: 1553-55-5) is formed when acetyl-CoA condenses with acetoacetyl-CoA in a reaction that is catalyzed by the enzyme HMG-CoA synthase in the mevalonate pathway or mevalonate-dependent (MAD) route, an important cellular metabolic pathway present in virtually all organisms. HMG-CoA reductase (EC 1.1.1.34) inhibitors, more commonly known as statins, are cholesterol-lowering drugs that have been widely used for many years to reduce the incidence of adverse cardiovascular events. HMG-CoA reductase catalyzes the rate-limiting step in the mevalonate pathway and these agents lower cholesterol by inhibiting its synthesis in the liver and in peripheral tissues. Androgen also stimulates lipogenesis in human prostate cancer cells directly by increasing transcription of the fatty acid synthase and HMG-CoA-reductase genes (PMID: 14689582 ). |
|---|
| Structure | C[C@](O)(CC(O)=O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C27H44N7O20P3S/c1-26(2,21(40)24(41)30-5-4-15(35)29-6-7-58-17(38)9-27(3,42)8-16(36)37)11-51-57(48,49)54-56(46,47)50-10-14-20(53-55(43,44)45)19(39)25(52-14)34-13-33-18-22(28)31-12-32-23(18)34/h12-14,19-21,25,39-40,42H,4-11H2,1-3H3,(H,29,35)(H,30,41)(H,36,37)(H,46,47)(H,48,49)(H2,28,31,32)(H2,43,44,45)/t14-,19-,20-,21+,25-,27+/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (3S)-3-Hydroxy-3-methylglutaryl-coenzyme A | ChEBI | | (S)-3-Hydroxy-3-methylglutaryl-CoA | ChEBI | | (S)-3-Hydroxy-3-methylglutaryl-coenzyme A | ChEBI | | HMG-CoA | ChEBI | | HMG-coenzyme A | ChEBI | | Hydroxymethylglutaroyl coenzyme A | ChEBI | | Hydroxymethylglutaryl-CoA | ChEBI | | (3S)-3-Hydroxy-3-methylglutaryl-CoA | HMDB | | (S)-3-Hydroxy-3-methylglutaryl coenzyme A | HMDB | | 3-Hydroxy-3-methylglutaryl CoA | HMDB | | 3-Hydroxy-3-methylglutaryl coenzyme A | HMDB | | Hydroxymethylglutaryl CoA | HMDB | | Hydroxymethylglutaryl coenzyme A | HMDB | | beta-Hydroxy-beta-methylglutaryl CoA | HMDB | | beta-Hydroxy-beta-methylglutaryl-CoA | HMDB | | beta-Hydroxy-beta-methylglutaryl-coenzyme A | HMDB | | Β-hydroxy-β-methylglutaryl CoA | HMDB | | Β-hydroxy-β-methylglutaryl-CoA | HMDB | | Β-hydroxy-β-methylglutaryl-coenzyme A | HMDB | | 3-Hydroxy-3-methylglutaryl-CoA | ChEBI, KEGG |
|
|---|
| Chemical Formula | C27H44N7O20P3S |
|---|
| Average Molecular Weight | 911.659 |
|---|
| Monoisotopic Molecular Weight | 911.157467109 |
|---|
| IUPAC Name | (3S)-5-[(2-{3-[(2R)-3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-3-hydroxy-3-methyl-5-oxopentanoic acid |
|---|
| Traditional Name | HMG-CoA |
|---|
| CAS Registry Number | 26926-09-0 |
|---|
| SMILES | C[C@](O)(CC(O)=O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C27H44N7O20P3S/c1-26(2,21(40)24(41)30-5-4-15(35)29-6-7-58-17(38)9-27(3,42)8-16(36)37)11-51-57(48,49)54-56(46,47)50-10-14-20(53-55(43,44)45)19(39)25(52-14)34-13-33-18-22(28)31-12-32-23(18)34/h12-14,19-21,25,39-40,42H,4-11H2,1-3H3,(H,29,35)(H,30,41)(H,36,37)(H,46,47)(H,48,49)(H2,28,31,32)(H2,43,44,45)/t14-,19-,20-,21+,25-,27+/m1/s1 |
|---|
| InChI Key | CABVTRNMFUVUDM-VRHQGPGLSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as (s)-3-hydroxy-3-alkylglutaryl coas. These are 3-hydroxy-3-alkylglutaryl-CoAs where the 3-hydroxy-3-alkylglutaryl component has (S)-configuration. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | (S)-3-hydroxy-3-alkylglutaryl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Alkyl phosphate
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Fatty amide
- Pyrimidine
- Phosphoric acid ester
- Tetrahydrofuran
- Tertiary alcohol
- Imidazole
- Heteroaromatic compound
- Azole
- Secondary alcohol
- Thiocarboxylic acid ester
- Amino acid
- Carboxamide group
- Amino acid or derivatives
- Carbothioic s-ester
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organosulfur compound
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesNot Available |
|---|
| Spectra |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 10V, Positive-QTOF | splash10-000l-0891000250-052d51ef28faa9ab5b73 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 20V, Positive-QTOF | splash10-000i-0971100000-dc16942c0a4531c8cb60 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 40V, Positive-QTOF | splash10-000i-1950000000-12dcb50a3caaf20a9aa5 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 10V, Negative-QTOF | splash10-00o4-4901031462-49fbf273ff8df18347bb | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 20V, Negative-QTOF | splash10-001i-3900110010-0be1f0212b5d814ea24c | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 40V, Negative-QTOF | splash10-057i-6900100000-fffa063c59d4fcd54e7a | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 10V, Negative-QTOF | splash10-03dm-0000000094-b818de142848216a6af3 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 20V, Negative-QTOF | splash10-0a4u-9500001521-8d0d8c41c4c284cc696f | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 40V, Negative-QTOF | splash10-004m-9202503430-001456d7b42e54584702 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 10V, Positive-QTOF | splash10-03di-0000000169-47d0ed82d5acca38b9e0 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 20V, Positive-QTOF | splash10-002r-1500000391-9d63a322b718612b8587 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxy-3-methylglutaryl-CoA 40V, Positive-QTOF | splash10-0a4i-0011900000-766a56d8ca4793434590 | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
- Mitochondria
- Endoplasmic reticulum
- Peroxisome
|
|---|
| Biospecimen Locations | Not Available |
|---|
| Tissue Locations | - Adrenal Gland
- Fibroblasts
- Intestine
|
|---|
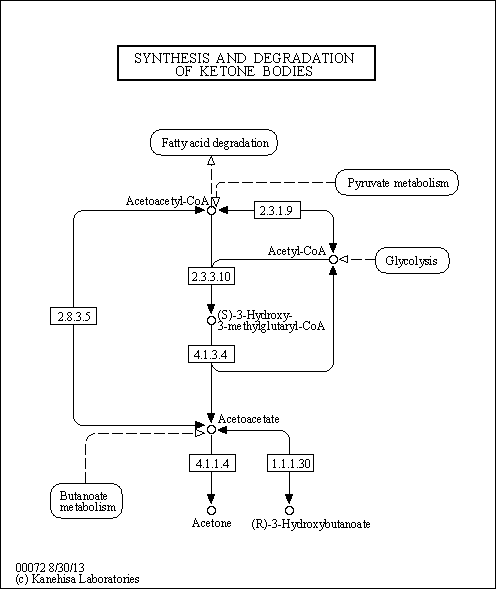
| Pathways | |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB030158 |
|---|
| KNApSAcK ID | C00007270 |
|---|
| Chemspider ID | 392859 |
|---|
| KEGG Compound ID | C00356 |
|---|
| BioCyc ID | 3-HYDROXY-3-METHYL-GLUTARYL-COA |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 445127 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15467 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | HMGCOA |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Williamson I P; Rodwell V W Isolation and purification of 3-hydroxy-3-methylglutaryl-coenzyme A by ion-exchange chromatography. Journal of lipid research (1981), 22(1), 184-7. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | - Sato T, Oouchi M, Nagakubo H, Chiba T, Ogawa S, Sato C, Sugimura K, Fukuda M: Effect of pravastatin on plasma ketone bodies in diabetics with hypercholesterolemia. Tohoku J Exp Med. 1998 May;185(1):25-9. [PubMed:9710942 ]
- Wysocki SJ, Wilkinson SP, Hahnel R, Wong CY, Panegyres PK: 3-Hydroxy-3-methylglutaric aciduria, combined with 3-methylglutaconic aciduria. Clin Chim Acta. 1976 Aug 2;70(3):399-406. [PubMed:947633 ]
- Lange Y, Ye J, Steck TL: Activation of membrane cholesterol by displacement from phospholipids. J Biol Chem. 2005 Oct 28;280(43):36126-31. Epub 2005 Aug 29. [PubMed:16129675 ]
- Li X, Liu L, Tupper JC, Bannerman DD, Winn RK, Sebti SM, Hamilton AD, Harlan JM: Inhibition of protein geranylgeranylation and RhoA/RhoA kinase pathway induces apoptosis in human endothelial cells. J Biol Chem. 2002 May 3;277(18):15309-16. Epub 2002 Feb 11. [PubMed:11839765 ]
- Mital BK, Garg SK: Anticarcinogenic, hypocholesterolemic, and antagonistic activities of Lactobacillus acidophilus. Crit Rev Microbiol. 1995;21(3):175-214. [PubMed:8845062 ]
- Nozaki S, Nakagawa T, Nakata A, Yamashita S, Kameda-Takemura K, Nakamura T, Keno Y, Tokunaga K, Matsuzawa Y: Effects of pravastatin on plasma and urinary mevalonate concentrations in subjects with familial hypercholesterolaemia: a comparison of morning and evening administration. Eur J Clin Pharmacol. 1996;49(5):361-4. [PubMed:8866629 ]
- Ubels FL, Muntinga JH, van Doormaal JJ, Reitsma WD, Smit AJ: Effects of initial and long-term lipid-lowering therapy on vascular wall characteristics. Atherosclerosis. 2001 Jan;154(1):155-61. [PubMed:11137095 ]
- Huang L, Wang Y, Grimm S: ATP-dependent transport of rosuvastatin in membrane vesicles expressing breast cancer resistance protein. Drug Metab Dispos. 2006 May;34(5):738-42. Epub 2006 Jan 13. [PubMed:16415124 ]
- Son BK, Kozaki K, Iijima K, Eto M, Kojima T, Ota H, Senda Y, Maemura K, Nakano T, Akishita M, Ouchi Y: Statins protect human aortic smooth muscle cells from inorganic phosphate-induced calcification by restoring Gas6-Axl survival pathway. Circ Res. 2006 Apr 28;98(8):1024-31. Epub 2006 Mar 23. [PubMed:16556867 ]
- Gianni L, Di Padova F, Zuin M, Podda M: Bile acid-induced inhibition of the lymphoproliferative response to phytohemagglutinin and pokeweed mitogen: an in vitro study. Gastroenterology. 1980 Feb;78(2):231-5. [PubMed:7350045 ]
- Jenke HS, Lowel M, Berndt J: Effect of alterations in vitro and in vivo of the cholesterol content in rat liver microsomes on the activity of 3-hydroxy-3-methylglutaryl-CoA reductase. Hoppe Seylers Z Physiol Chem. 1983 Feb;364(2):135-40. [PubMed:6840702 ]
- Naseem SM, Heald FP: Sex mediated lipid metabolism in human aortic smooth muscle cells. Biochem Biophys Res Commun. 1987 Apr 14;144(1):284-91. [PubMed:3579907 ]
- Knight BL, Patel DD, Soutar AK: The regulation of 3-hydroxy-3-methylglutaryl-CoA reductase activity, cholesterol esterification and the expression of low-density lipoprotein receptors in cultured monocyte-derived macrophages. Biochem J. 1983 Feb 15;210(2):523-32. [PubMed:6305342 ]
- Martin J, Denver R, Bailey M, Krum H: In vitro inhibitory effects of atorvastatin on cardiac fibroblasts: implications for ventricular remodelling. Clin Exp Pharmacol Physiol. 2005 Sep;32(9):697-701. [PubMed:16173924 ]
- Plotkin D, Miller S, Nakajima S, Peskin E, Burkman R, Richardson D, Mitchel Y, Waldstreicher J, Liu M, Shapiro D, Santoro N: Lowering low density lipoprotein cholesterol with simvastatin, a hydroxy-3-methylglutaryl-coenzyme a reductase inhibitor, does not affect luteal function in premenopausal women. J Clin Endocrinol Metab. 2002 Jul;87(7):3155-61. [PubMed:12107216 ]
- Gil G, Smith JR, Goldstein JL, Brown MS: Optional exon in the 5'-untranslated region of 3-hydroxy-3-methylglutaryl coenzyme A synthase gene: conserved sequence and splicing pattern in humans and hamsters. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1863-6. [PubMed:3470763 ]
- Freeman MR, Solomon KR: Cholesterol and prostate cancer. J Cell Biochem. 2004 Jan 1;91(1):54-69. [PubMed:14689582 ]
|
|---|